
Newsroom
Revolutionary CuRVE Technology for Rapid Protein Labeling in 3D Tissue Analysis
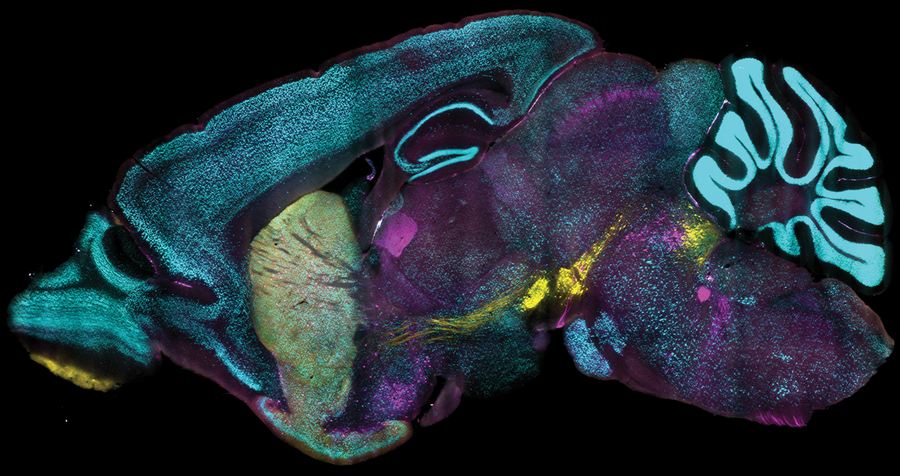
A groundbreaking technology developed at MIT is revolutionizing the way scientists label proteins in millions of individual cells within fully intact 3D tissues. This innovative approach allows for rapid, uniform, and versatile protein labeling, enabling researchers to process large tissue samples in just one day. The recent study published in Nature Biotechnology highlights how this method can uncover valuable insights that conventional labeling techniques often miss.
The Importance of Protein Profiling in Research
Profiling the proteins produced by cells is a fundamental aspect of biological and neuroscience research. The proteins expressed by a cell at any given time can indicate its functions and responses to various conditions, including diseases and treatments. Despite advancements in microscopy and labeling technologies, researchers have faced challenges in accurately tracking protein expression across densely packed cells in intact, whole tissues. Traditional methods often involve slicing tissues into thin sections, limiting the ability to analyze cellular protein expression in a comprehensive manner.
Overcoming Technical Challenges in Tissue Labeling
Investigating cellular molecules typically requires the dissociation of tissues into single cells or the creation of thin tissue slices, as light and chemicals cannot penetrate deeply into denser tissues. Previous innovations like CLARITY and SHIELD have enabled whole-organ investigations by making tissues transparent. However, a reliable method for chemically labeling entire organs was still needed to gain meaningful scientific insights.
The newly developed method, known as CuRVE, marks a significant milestone in addressing this challenge. It represents a fresh approach to uniformly processing large and dense tissue samples. The researchers detail how they overcame technical barriers through an implementation called eFLASH, showcasing its effectiveness with compelling demonstrations that led to new discoveries in neuroscience.
Advancements in Protein Labeling Chemistry
According to Dae Hee Yun, a co-lead author and senior application engineer at LifeCanvas Technologies, this represents a substantial advancement in the performance of protein labeling technology. The study also includes contributions from Young-Gyun Park, a former MIT postdoc now serving as an assistant professor at KAIST in South Korea.
“The CuRVE strategy effectively resolves the speed mismatch of antibody diffusion and binding, enabling faster and more uniform labeling of large tissue samples.”
The slow diffusion of antibodies into large 3D tissue samples poses a challenge for uniform labeling. Antibodies quickly bind to target proteins but penetrate tissues slowly, resulting in uneven labeling across the sample. The CuRVE strategy effectively resolves this speed mismatch by continuously controlling antibody binding rates while enhancing antibody permeation throughout the tissue. The researchers utilized advanced computational simulations to optimize their approach, adjusting parameters such as binding rates and tissue densities.

Implementation and Comparative Results
The team built upon a previous technology called SWITCH, which allowed for temporary control over antibody binding. By screening for more suitable chemicals, they discovered deoxycholic acid could effectively modulate binding speed and improve labeling outcomes. This eFLASH system significantly accelerated antibody movement through tissues using stochastic electrotransport technology.
The researchers compared eFLASH labeling with conventional genetic engineering methods that cause cells to fluoresce when certain proteins are being expressed. While genetic methods do not require antibody dispersion, they may not accurately reflect actual protein presence. The study demonstrated discrepancies between antibody labeling and genetic reporting, highlighting the importance of integrating both methods for a more comprehensive understanding of protein expression.
Conclusion and Acknowledgments
This innovative approach not only facilitates faster labeling but also enhances the quality of data gathered from protein profiling. The authors emphasize that combining antibody labeling with genetic methods provides a richer context for understanding cellular functions and variations.
This research was supported by various organizations, including the Burroughs Wellcome Fund, the Searle Scholars Program, and the National Institutes of Health.



