
Newsroom
Revolutionizing Drug Discovery with AI: Efficiently Synthesizable Molecule Design
Artificial intelligence revolutionizes drug discovery by generating synthesizable molecular structures quickly, enabling faster development of effective medicines that can be realistically produced in the laboratory.
Innovating Molecular Design with AI
Pharmaceutical companies are increasingly adopting artificial intelligence to streamline the discovery of new medicines. Traditional machine-learning models propose molecules with potential disease-fighting properties but often recommend structures that are impossible to synthesize. This challenge limits their practical application since unproducible molecules cannot be experimentally tested.
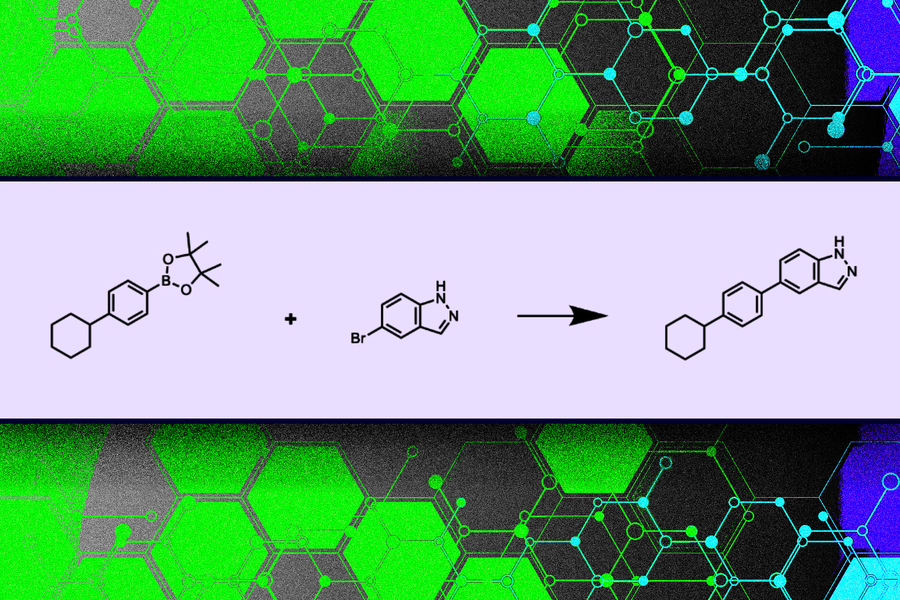
A Synthesis-Constrained Molecular Builder Model
Researchers at MIT developed a novel model that guarantees the generation of chemical molecules which can be synthesized. Unlike atom-by-atom methods, their approach constructs molecules block by block using purchasable building blocks and valid chemical reaction templates. This strategy reduces the search space and ensures that each proposed molecular structure follows chemical laws and is practically manufacturable.
The model outputs both the final molecular structure and the synthesis tree outlining how chemicals and reactions combine step-by-step. Training involved feeding the system complete molecules along with their synthesis paths so it could learn to recreate these sequences autonomously.
Optimization and Practical Applications
Once trained, the model can optimize molecules to meet specific properties while ensuring synthesizability. It reconstructs over half of tested molecules within seconds and suggests structures with stronger binding affinities compared to existing methods. For example, it proposed candidates potentially better at binding SARS-Cov-2 proteins than current inhibitors, though these results await experimental confirmation.
“Instead of designing molecules atom by atom, building them block by block and reaction by reaction transforms drug discovery into an efficient, synthesizable process.”
Future Directions in Computer-Aided Synthesis
The team plans to enhance their chemical reaction templates to further improve model performance and broaden applicability to diverse disease targets. This method represents a significant advance towards automated molecule design combined with synthesis planning, inspiring future research and accelerating pharmaceutical innovation worldwide.



